Explore our expert-made templates & start with the right one for you.
An Easier Way to Build a Data Ingestion Pipeline for Batch and Streaming Data
How to Avoid Big Data “Indigestion”
Ingesting data into your pipeline architecture is a critical first step in any data engineering project. While seemingly simple, there are numerous challenges that organizations face when trying to build out their ingestion architectures:
- Data comes from a variety of sources, both streaming and batch, which must be ingested into a common platform.
- The data itself can be in many different formats, including but not limited to JSON, CSV, TSV, Parquet, AVRO, ORC, and so on. Data may also be compressed using several different algorithms.
- Ingested data does not always come with a known and static schema. When schemas are modified over time (schema evolution), ingestion pipelines can break.
- Scaling data ingestion can be difficult to implement and manage, especially for event data where volumes can vary substantially, leading to performance or cost issues.
Typically architects have avoided this data “indigestion” in a variety of ways:
- They can code a pipeline manually via technologies such as Spark, Python, and Airflow. This method is powerful and versatile. But it is also time-intensive and error-prone, and finding data engineers with the requisite knowledge and experience is both difficult and expensive.
- They can use a managed Spark service to take some of the labor out of programming their own pipelines. Most of the leading data engineering providers – Amazon Web Services, Databricks, Microsoft Azure, and Google Cloud – offer such a service. But these work best in homogeneous environments, in which you give over your full stack (not just your ingestion pipelines) to the same vendor’s technologies, and essentially lock yourself out of new tool and technology developments that are external to your vendor’s “walled garden.”
- They can rely on connector software such as Fivetran, Airbyte, or a number of other offerings. These remove some of the hazards of manual pipeline construction, such as keeping up with changes to source system APIs. But they may not easily scale or handle elastic workloads efficiently, especially with regard to streaming data.
A Declarative Pipeline Platform for Batch and Streaming Data
You can also take a declarative approach to creating ingestion pipelines. In this approach, you define your ingestion parameters and instantiate your pipelines using only declarative SQL; no knowledge of Spark, Scala, or other complex languages is necessary.
Here’s an example, using Upsolver SQLake.
SQLake is a data pipeline platform that uses a declarative approach to specifying pipelines. This means you define your sources, target, and transformation outputs, and it optimizes the execution automatically, based on your definitions. SQLake handles both streaming and batch data ingestion at scale, using a simple SQL syntax to define operations.
There are 3 primary components:
- A connection to the system currently holding the data.
- A staging table where raw data is loaded for downstream processing.
- A job that runs continuously, copying data from the source connection into the staging table.
Here are some details on each of these 3 components.
Connections in data ingestion pipelines
Connections define the details needed to connect to and authenticate your data sources. SQLake provides connectivity to 3 types of data sources:
- Amazon S3
- Amazon Kinesis
- Apache Kafka
All connections are defined similarly, using SQL, with connection-specific parameters for authentication. Below are example S3 and Kinesis connections; sensitive information is redacted. For full connection syntax, please refer to the SQLake documentation.
CREATE S3 CONNECTION upsolver_s3 AWS_ROLE = '<arn:aws:iam::########:role/<role>' EXTERNAL_ID = 'SAMPLES' READ_ONLY = TRUE COMMENT = 'S3 Data provided by Upsolver';
CREATE KINESIS CONNECTION upsolver_sa_kinesis AWS_ROLE = ‘<arn:aws:iam::########:role/<role>' EXTERNAL_ID = '<EXTERNAL_ID>' REGION = 'us-east-2' READ_ONLY = true;
To ingest data in AWS you also need a connection to an AWS Glue Catalog. The initial Upsolver SQLake installation includes a default Glue Catalog. But you can create additional Glue Catalog connections as needed.
After connections are successfully created, they display in your catalog view and are available for you to browse:

Staging Tables in Data Ingestion Pipelines
After you’ve defined a connection to the data source, you specify where to land the data. SQLake ingests data as Apache Parquet files in a managed S3 bucket. It creates a metastore catalog so that ingested data can be queried, verified, and made available for transformations. You can also define your staging tables with partitioning for performance at scale.
Here is the simplest form of a staging table. It consists only of a defined name and a partition key that maps to the date the event data was pulled from the source.
CREATE TABLE default_glue_catalog.<DB NAME>.<TABLE_NAME>() partition by $event_date;
There’s no need to define columns when the staging table is created; SQLake dynamically detects the schema on read and maps these fields during data ingestion, including nested data in the source as arrays.
Jobs in Data Ingestion Pipelines
The third step in an ingestion pipeline is to create a job that continually moves data from the source connection into your staging table. Below you can view the syntax for this kind of job; most parameters are set to default for simplicity. In many cases, this simple default job is sufficient, as with this S3 example.
CREATE JOB load_orders_raw_data_from_s3
START_FROM = BEGINNING
CONTENT_TYPE = AUTO
DATE_PATTERN = 'yyyy/MM/dd/HH/mm'
AS COPY FROM S3 upsolver_s3_samples BUCKET = 'upsolver-samples' PREFIX = 'orders/'
INTO default_glue_catalog.database_9c2e45.orders_raw_data;
Below are the most important job parameters; find the full list of job parameters in the SQLake documentation.
START_FROM = NOW | BEGINNING
This tells the ingestion job whether to start ingesting new files (NOW) or to go back and begin reading in files from as far back as they go (BEGINNING). In this way you can decide whether to include historical data ingested into your staging tables.
DATE_PATTERN = 'yyyy/MM/dd/HH/mm'
This tells the ingestion job the date pattern of your S3 partition structure. In the example above, files are partitioned into folders matching yyyy (year), MM (month), dd (day), HH (hour), and mm (minute).
CONTENT_TYPE = AUTO | JSON | CSV | AVRO | PARQUET | ORC…
If you choose AUTO, SQLake attempts to determine the content type of the files. This is often the best option, though if you are certain your files are of a specific type you can define it here.
COMPRESSION = AUTO | GZIP | SNAPPY | …
If your source data might be compressed, setting COMPRESSION to AUTO tells SQLake to try detecting and uncompressing the source data. This is especially helpful if you aren’t sure which compression algorithms to use, as SQLake automatically determines the compression method and then applies the corresponding decompression routines.
FILE_PATTERN = ‘<pattern>’
The default is to load in all files contained within the specified buckets. However, if you want only certain files, you can include a regex filter.
For jobs that read off of a streaming source (such as Amazon Kinesis or Apache Kafka), you can define several additional parameters.
READER_SHARDS = <integer>
This enables SQLake to scale up the number of readers used to ingest your stream. For small-to-medium streams, a single shard should suffice. But if your stream is processing more than 2 MB/second of peak streaming volume, then for every 2 MB beyond that you can increment this value by 1. For example, if your peak streaming volume is 8 MB/s, set READER_SHARDS to a value of 4.
For all ingestion types, SQLake uses an auto-sharding architecture that scales up the number of parallel processes used to ingest data.
Monitoring and Tracking Your Declarative Ingestion Pipeline
You’ve created a connection to your source data, built a staging table into which to ingest the data, and defined a COPY FROM job to ingest from source to staging. With your ingestion job now running, it’s time to monitor and track its progress. There are a variety of ways you can do this.
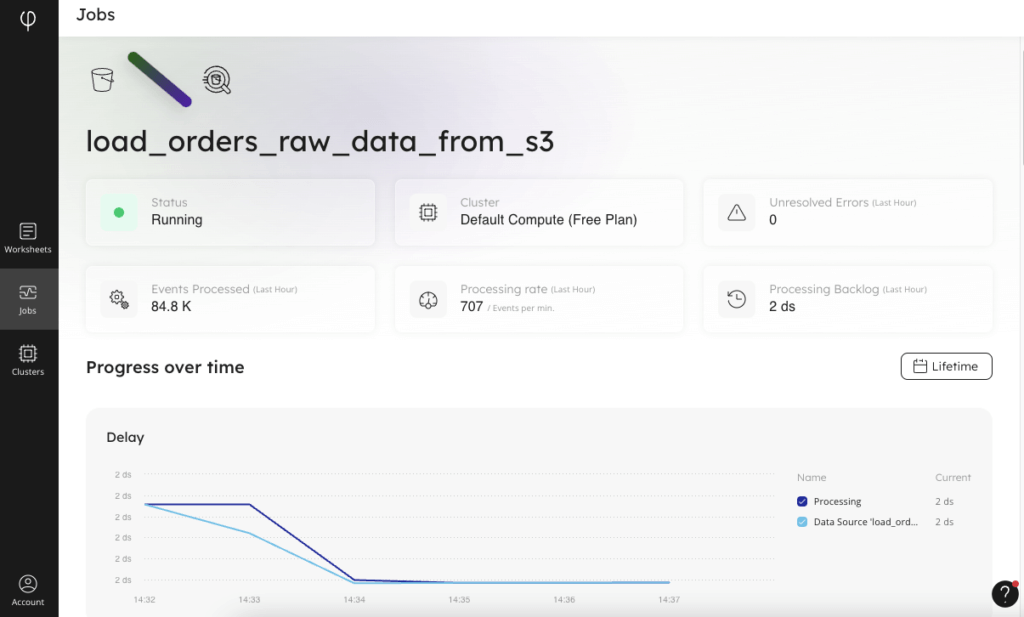
Visually, there is a “Jobs” interface in SQLake that displays the status of each job, with high-level information about how the job is running and how much data is being processed over time. In addition to the graphical job monitoring interface, there are system tables in your catalog view that you can query to examine the health of the various components of each job.
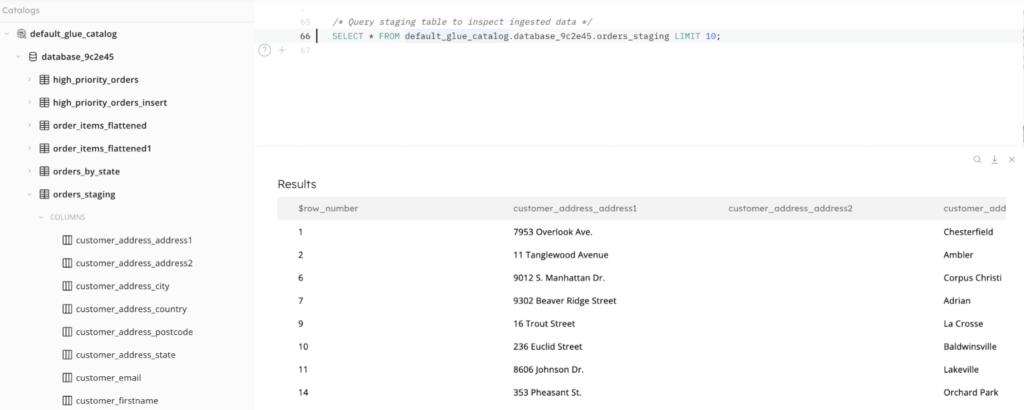
And you can go beyond monitoring the job to querying the staging tables. Because SQLake creates metadata catalog entries for your staging tables and partitions the incoming data, it can be efficiently queried from within a SQLake worksheet. Inspecting the data you’ve ingested is as simple as writing a SELECT statement against the staging table, as you can see in the screenshot below.
Summary: Building a Declarative Data Ingestion Pipeline
To help you ingest a mix of batch and streaming data, SQLake connects to a variety of data sources, using a common framework and familiar SQL syntax. You can ingest batch and streaming data in parallel, into a standardized Parquet format, and then make it available for downstream transformations and output.
Automated schema, format, and compression detection and conversion. If your data sources include a variety of data formats (JSON, CSV, AVRO, ORC, and so on) and a mix of compressed and uncompressed formats, SQLake automatically identifies and decompresses data as it is ingested.
Low-latency ingestion at scale. Due to its use of compression, compaction, partitioning, and dynamic scaling of parallel resources, SQLake has been shown to ingest extremely large volumes of data efficiently and with up-to-the-minute latency. See a benchmark we performed recently using the Upsolver engine that powers SQLake.
Ingestion observability. Both during and after ingestion, SQLake includes built-in observability of ingestion job performance, as well as simple query capabilities to inspect the incoming data. Using Athena queries from within a worksheet, you can verify that the ingested data is what you expect it to be.
SQLake worksheet:
/* This template walks you through ingesting S3 data and staging it in a table that can be queried. 1. Create a connection to S3 providing the IAM credentials needed to access data. 2. Create a staging table to hold the raw data ingested from the S3 source. 3. Create a job to ingest the data from S3 into the staging table created in the previous step. 4. Query your staging table to verify data ingestion /* 1. Create an S3 connection to manage IAM credentials for jobs that need to access data in S3 */ CREATE S3 CONNECTION upsolver_s3_samples AWS_ROLE = 'arn:aws:iam::949275490180:role/upsolver_samples_role' EXTERNAL_ID = 'SAMPLES' READ_ONLY = TRUE; /* 2. Create an empty table to use as staging for the raw data. */ CREATE TABLE default_glue_catalog.database_c10c46.orders_raw_data() PARTITIONED BY $event_date; /* 3. Ingest raw data from your bucket into the staging table */ CREATE JOB load_orders_raw_data_from_s3_2 CONTENT_TYPE = JSON AS COPY FROM S3 upsolver_s3_samples BUCKET = 'upsolver-samples' PREFIX = 'orders/' INTO default_glue_catalog.database_c10c46.orders_raw_data; /* 4. Query your raw data in the staging table Note: It may take 3-4 minutes for the data to appear in your staging table. */ SELECT * FROM default_glue_catalog.database_c10c46.orders_raw_data limit 10; /* OPTIONAL You can gauge progress of the job by inspecting the SystemTable. The following query will return results when the job begins to process data and write the results to S3. */ SELECT job_name, stage_name, task_start_processing_time, task_end_processing_time, bytes_read, task_error_message FROM SystemTables.logs.task_executions WHERE job_name = 'load_orders_raw_data_from_s3' AND (stage_name = 'write to storage' OR stage_name = 'parse data') ORDER BY task_start_processing_time DESC LIMIT 10;
Try SQLake for Free
SQLake is Upsolver’s newest offering. It lets you build and run reliable data pipelines on streaming and batch data via an all-SQL experience. Try it for free for 30 days. No credit card required.

